Article de blogue en français ici
Borealis upgraded to Dataverse version 5.13 earlier this year (see previous blog post). The Borealis team has been configuring additional new features that bring support for:
To test these features, feel free to use our demo sandbox environment. Please reach out if you have any questions or feedback.
Github integration: Upload to Borealis using a GitHub Action
What is it?
GitHub repository content can be uploaded to an existing Borealis dataset using a “GitHub Action” called the Dataverse Uploader Action.
What is the use-case?
This integration provides a simplified way to backup your GitHub repository to a Borealis dataset. This customizable action can allow you to:
- Upload the entire GitHub repository or specific subdirectories
- Automatically synchronize at trigger events (e.g., push, release) or manually using the workflow dispatch event
- Turn on/off deleting dataset content before uploading from GitHub
- Turn on/off automatically publishing a new version of the dataset
How do I access this feature?
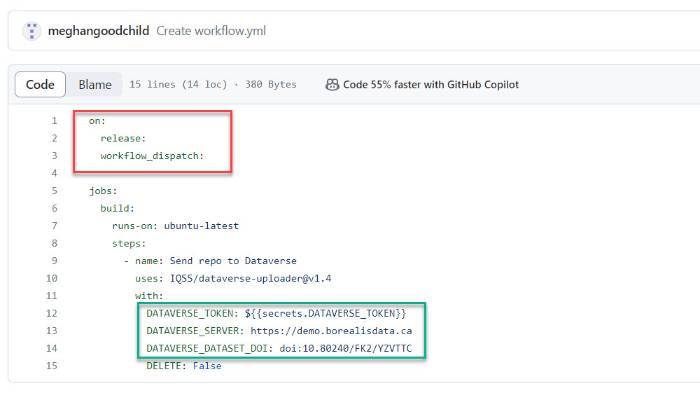
- Within your GitHub repository, create a YML file (i.e., workflow.yml) file within the directory called .github.workflows/ (see screenshot below)
- Enter your configurations, such as the trigger events (shown below in top box, lines 1 to 3), and whether you would like to upload the entire repository (default) or specific subdirectories.
- Add your API token as a secret variable (shown below in bottom box, line 12)
- Input the server URL (e.g., https://borealisdata.ca, https://demo.borealisdata.ca) and dataset DOI (e.g., doi:xx.xxxx/xxx/xxxxxx) (shown below in bottom box, lines 13 and 14)
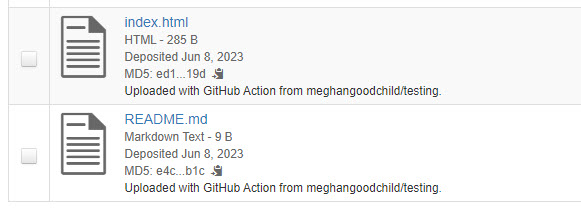
After the GitHub action uploads the files to Borealis, you will see the files show up in the file list with file-level metadata “Uploaded with GitHub Action from” with the name of your repository appended (see screenshot below).
ZIP previewer and option to download a single file within a zip
What is it?
ZIP archives can now be previewed within the browser and individual files can be selected for download.
What is the use-case?
Originally users needed to download an entire ZIP archive in order to view the files inside, even if they only needed a subset of the files. This new previewer allows users to review the files prior to download and only download a subset as needed.
How do I access this feature?
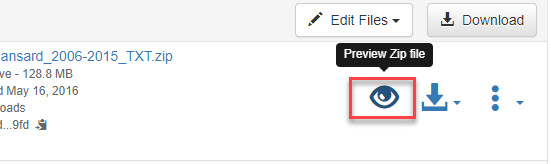
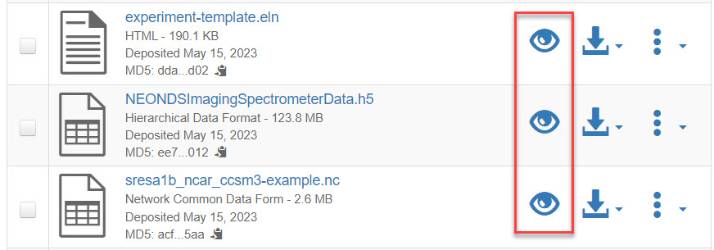
- Navigate to a ZIP archive and click on the preview icon (see screenshot).
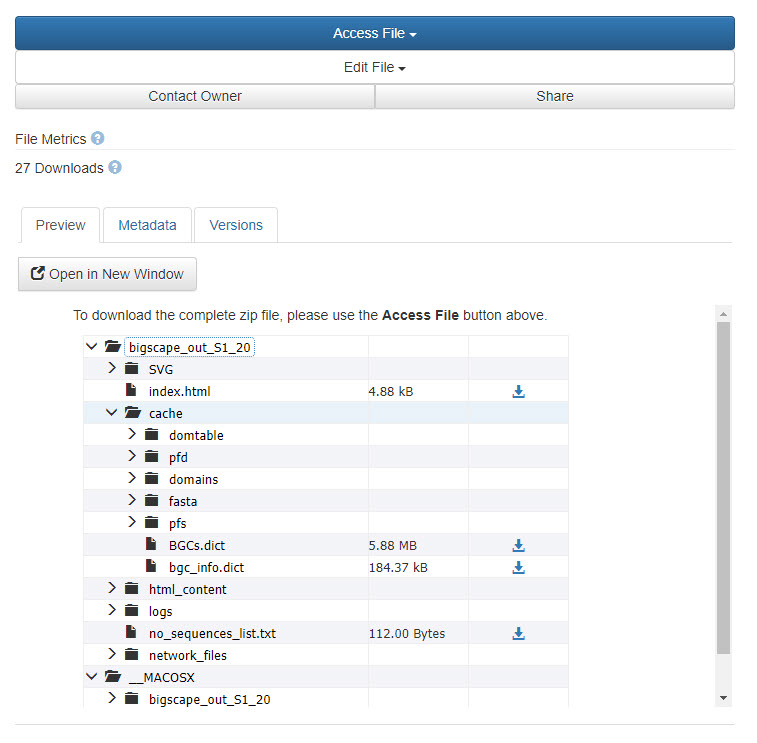
2. After accepting any access terms of use, you will be able to view the structure of the ZIP archive, navigate the file hierarchy, and download individual files (see screenshot below).
NcML and ELN (NcML) previewer
What is it?
With the latest upgrade, the Dataverse software now detects NetCDF and HDF5 files based on their content and attempts to extract metadata in NcML (XML) format and save as an auxiliary files. Additionally, ELN files are also detected.
We’ve now configured previewers to allow users to preview NcML files and ELN files. For more information, see our previous blog post and the Advanced Guide.
What is the use-case?
NetCDF (Network Common Data Form) is a machine independent data format, an international standard of the Open Geospatial Consortium, and commonly used in environmental and climate sciences.
HDF5 (Hierarchical Data Format) is an open source file format that supports large and complex data.
The ELN file format was developed to improve interoperability among different ELN software. This archive format allows for the import/export of research data, such as experimental results, protocols, descriptions, templates, etc.
These previewers allow users to explore these files before downloading.
How do I access this feature?
Navigate to the file list for files that have been detected as NetCDF, HDF5, or ELN. Click on the eye icon to access the previewer.
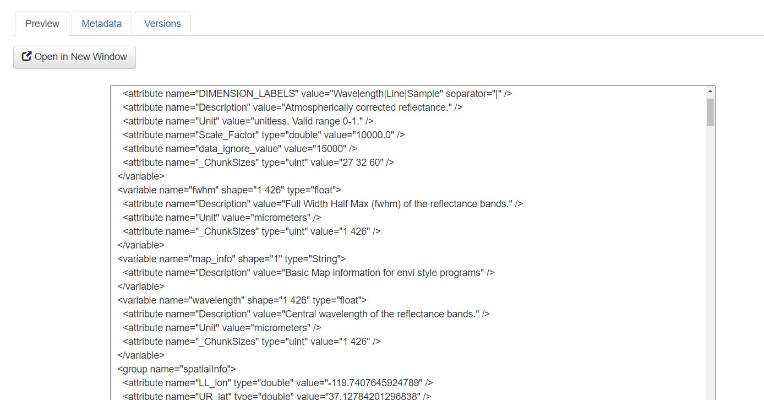
Once the previewer opens, users will be able to view the contents of the file with the NetCDF previewer.
Computational Workflow Metadata Block
What is it?
A computational workflow describes a process to coordinate multiple computational tasks and their data dependencies that lead to the resulting finalized dataset, for example: running code, using command-line tools, accessing a database, submitting a job to a compute cloud resource, and execution of data processing scripts. In the diagram below, an example workflow is shown where the original data follows a series of tasks and conditions that are mapped out as steps, ultimately resulting in the final data product.
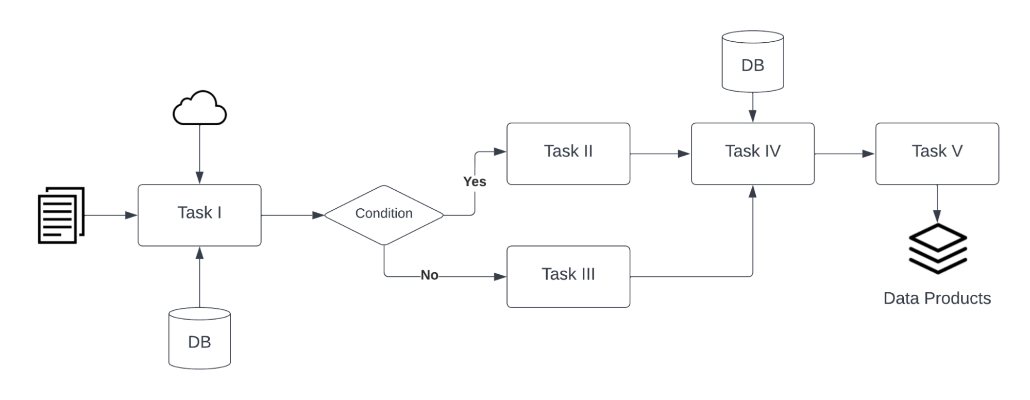
(Source: Institute for Quantitative Social Science. (accessed 2023-06-01). Dataverse User Guide https://guides.dataverse.org/en/5.13/user/dataset-management.html#computational-workflow)
Computational workflow information can be included within Borealis in two ways. First, users can apply a “workflow” tag to computational workflow files that are uploaded to Borealis. Second, Borealis now has a computational metadata workflow metadata block that can link to an external code repository where the related code and workflow steps are stored. These two options are described further below.
What is the use-case?
Including computational workflows as part of dataset documentation is increasingly becoming part of best practices to support transparent data management and reproducibility. Computational workflow information helps future dataset users better understand how the original data are transformed into the finished product.
How do I access this feature?
There are two options for including computational workflow information within Borealis:
- Create a computational workflow file using a framework or tool (e.g., Common Workflow Language (CWL), R Notebook, workflow registries). Then, upload your file to your dataset and add a custom tag called “workflow” (see screenshot below).
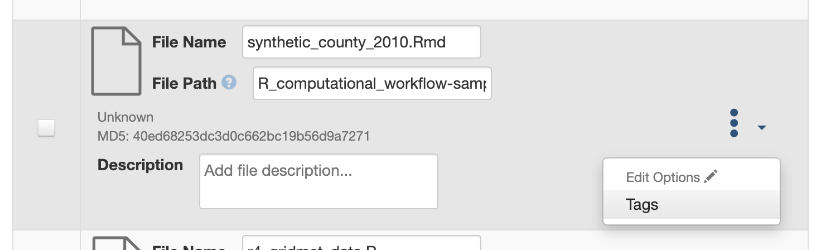
(Source: Institute for Quantitative Social Science. (accessed 2023-06-01). Dataverse User Guide https://guides.dataverse.org/en/5.13/user/dataset-management.html#computational-workflow)
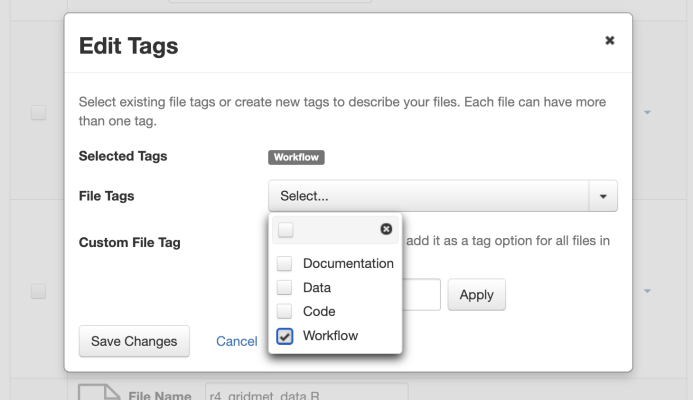
(Source: Institute for Quantitative Social Science. (accessed 2023-06-01). Dataverse User Guide https://guides.dataverse.org/en/5.13/user/dataset-management.html#computational-workflow)
2. Once the dataset is saved, go to the “Metadata” tab and select “Add + Edit Metadata.” Navigate to the computational workflow metadata block and add details to link to external code repositories that contain the code and related details about computational workflow steps. The fields can contain details about the type of computational workflow framework, the external code repository URL where the related code is located, and the URL to documentation or text describing the Computational Workflow and its use.
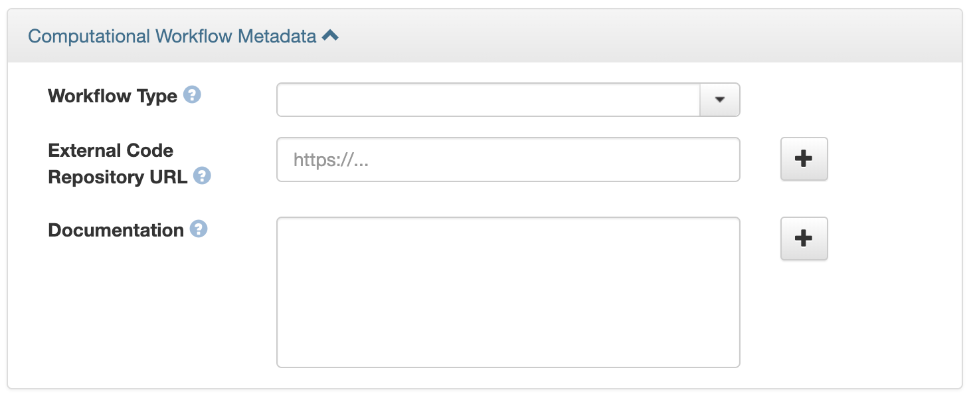
For more information:
Goble et al., (2020) FAIR Computational Workflows. https://direct.mit.edu/dint/article/2/1-2/108/10003/FAIR-Computational-Workflows
Dataverse Guides. “Dataset + File Management.” https://guides.dataverse.org/en/5.13/user/dataset-management.html#computational-workflow